IRIDA 20.05 Released
May 28, 2020
IRIDA’s second release for 2020, version 20.05, is ready for download. Find the new release on our GitHub at https://github.com/phac-nml/irida/releases.
This release includes some significant features to help IRIDA users take advantage of the changing landscape for sequencing and analysis tools used in today’s public health laboratories. It also has continued the trend of refreshing a number of regularly used pages and libraries to help the IRIDA team to be able to continue building new features for future releases.
Uploaded Assembly Support
This version of IRIDA has added the ability for users and tool developers to upload assemblies to IRIDA. Previous versions of IRIDA relied on assemblies generated by one of IRIDA’s assembly pipelines. Users may upload assemblies through the sample page similar to other sequence files, where they will be added alongside assemblies generated within IRIDA.
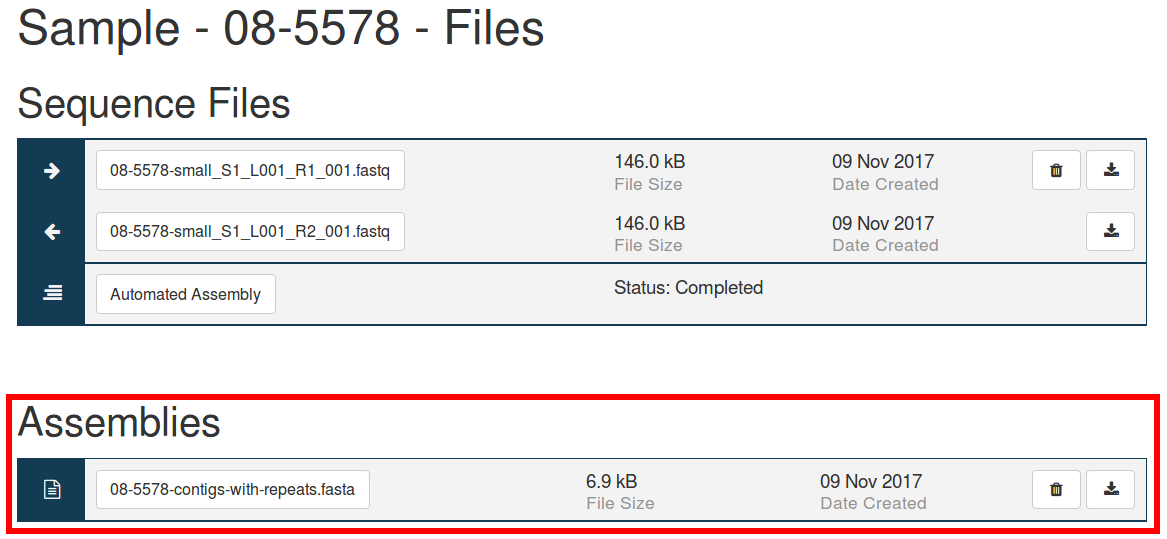
Developers are now able to write tools to upload and read assemblies through IRIDA’s REST API through newly added API endpoints. The IRIDA uploader does not yet support uploading assemblies, but watch for updates to that project enabling this support.
This version of IRIDA also adds support for exporting assemblies to Galaxy through our Galaxy Importer project, and to the command line through the NGS Archive Linker. IRIDA’s analysis pipelines currently do not support running tools with assembly files as input, but this feature is expected in future IRIDA versions.
FAST5 File Support
IRIDA 20.05 adds preliminary FAST5 file format support. Similar to the assemblies feature above, these files can be uploaded through the samples page and endpoints are available to developers to upload via the REST API. FAST5 files can currently be in one of two formats:
- Single
.fast5files - a combined sequence file that contains reads for a single sample. .fast5.tar.gzfiles - A gzipped directory of FAST5 files for a single sample.
In this initial release, FAST5 files can be exported to Galaxy through our Galaxy Importer project,and to the command line through the NGS Archive Linker. IRIDA’s analysis pipelines currently do not support FAST5 data, but this will be reviewed for future versions.
FAST5 support should be considered a beta feature, and is expected to change with future releases. These data formats will be reviewed in future IRIDA versions and support may be added or removed as necessary.
Analysis Result Page Refresh
IRIDA’s analysis results pages have been completely rewritten to help support future analysis pipeline development. This refresh has moved IRIDA’s analysis results pages to use the React framework to help improve future development of analysis visualizations and results tools.
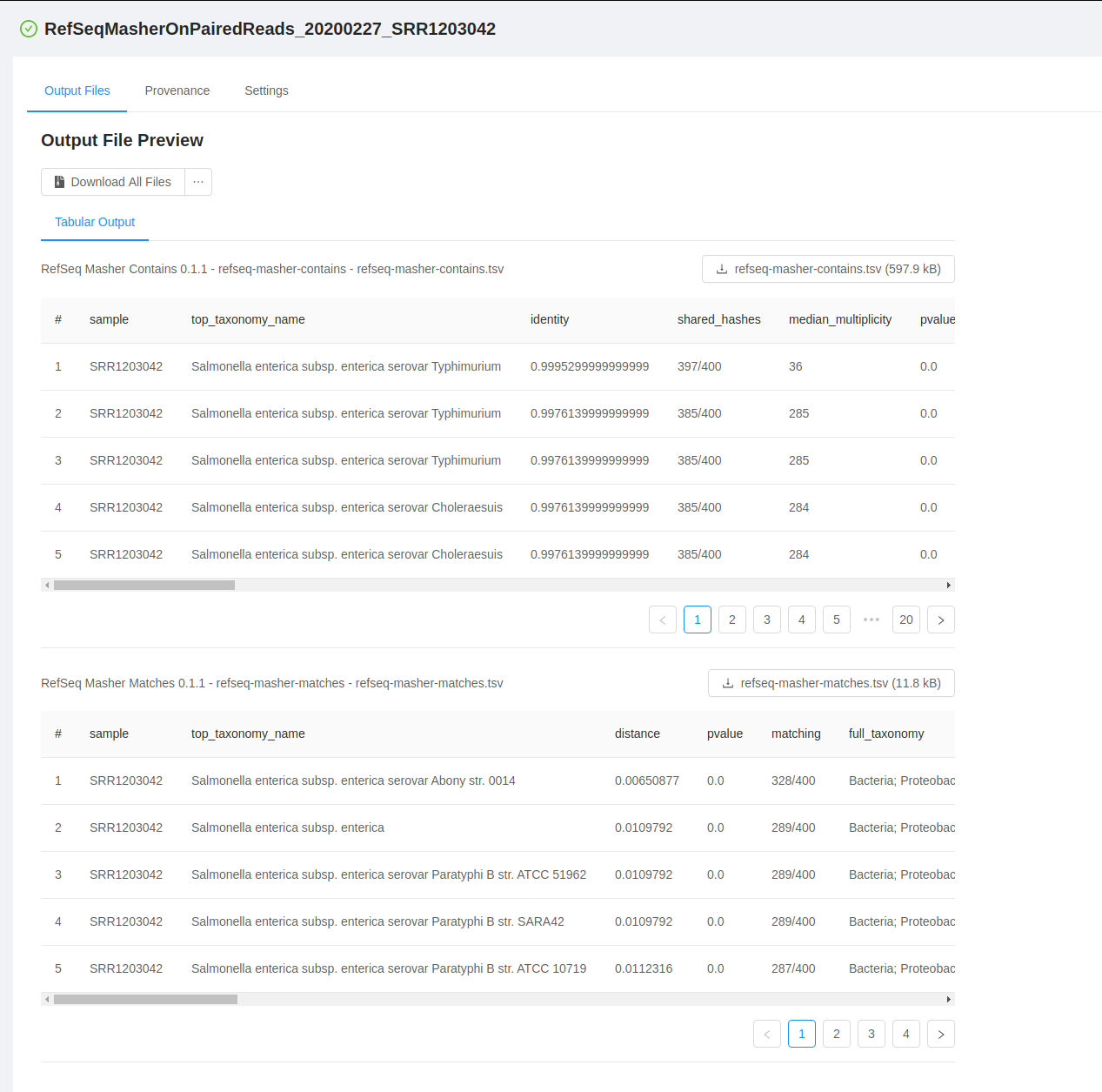
These results pages include built in viewers for specialized workflows like SNVPhyl and SISTR. Plugin developers can take advantage of default viewers for a number of file formats including JSON, Newick tree files, tabular data (csv, tdf, Excel), image files, and text-based file formats.
Customizable Login Page
IRIDA’s login page can now be customized by institutions to add their own logos, branding, or information needed for users logging in to IRIDA at their institution. A default login page is included. See more about customizing IRIDA for your institution in our documentation.
Other changes
- Updated several parts of the IRIDA UI to use modern styling, JS, and UI components including:
- Client listing page
- Announcements page
- Project breadcrumbs
- Remote API listing page
- Users listing page
- User groups pages
- Project members page.
- Updated FastQC in IRIDA to version 0.11.9.
For a full list of changes in this IRIDA release, see our changelog at https://github.com/phac-nml/irida/blob/20.05/CHANGELOG.md.