Platform Overview
The Goal of IRIDA
The Integrated Rapid Infectious Disease Analysis (IRIDA) platform is a user-friendly, distributed, open source bioinformatics and analytical web platform, developed to support real-time infectious disease outbreak investigations using whole genome sequencing data. IRIDA is designed for use in public health, food safety, and clinical microbiology labs, and is also suitable for general infectious disease comparative microbial genomics. The platform is scalable and is currently transforming the Canadian public health ecosystem.
While IRIDA was initially created to support the Canadian public health system, instances can be independently installed on local computing resources, enabling private and secure analyses according to organizational policies and governance. IRIDA supports controlled, collaborative data sharing and data analysis for users on its platform and for users on other IRIDA platforms. IRIDA’s data management capabilities enable secure upload, storage and management of all sequences and metadata, and provide data provenance and auditability as required by clinical laboratories. Easy-to-run pipelines make sophisticated genomics analyses accessible to lab analysts, while providing power-users with modularity and customizability.
Currently, IRIDA offers:
- End-to-end management of genomic sequence data and metadata
- High performance computing and analysis of genomic data
- cgMLST analyses
- Salmonella in silico serotyping
- Informative visualizations of genomic analysis results
- Controlled data access
- REST API for interacting with external programs
- Data sharing between instances
- Industry standard data security controls
- Data standardization enabling value-added activities
- User support
Also:
- IRIDA software is FREE
- IRIDA software is open source (under Apache 2.0 license)
- VMs and a public instance are available for user evaluation. Please contact irida-mail@sfu.ca if you would like to use it.
- Development reflects best clinical, bioinformatics, data stewardship practices
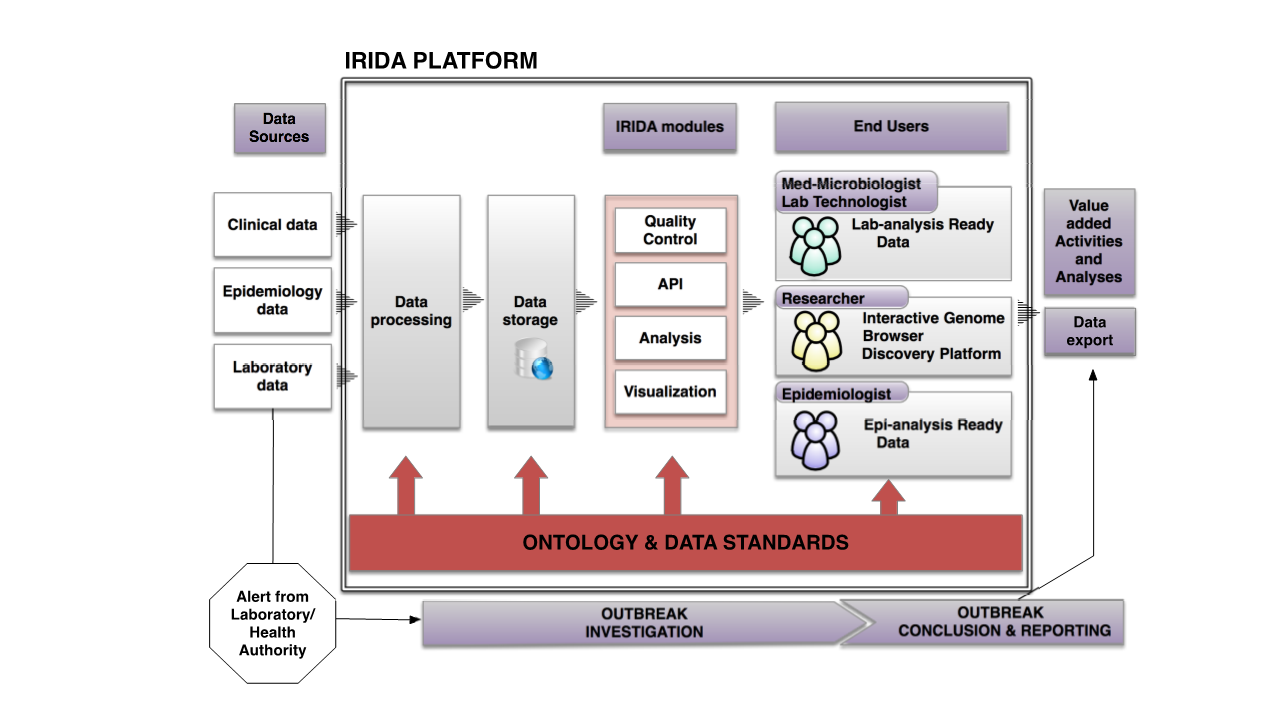
IRIDA strives to shield public health, regulatory and clinical microbiology laboratories from the computational challenges of genomics big data by providing solutions for data storage and management, analyses, and data sharing, to better incorporate WGS technology into routine operations.