Workflows and Analyses
IRIDA’s Core Functionality
- Secure end-to-end data management
- Fit-for-purpose analyses
- Data sharing
- Data visualization
- Enhanced Data Harmonization and Integration
Data Management
IRIDA instances behave like secure, independent, data management environments, allowing users to maintain complete control over their data during upload, analyses and storage. Through IRIDA’s distributed framework, data sharing between IRIDA instances can be enabled by altering access permissions on samples, projects and analyses.
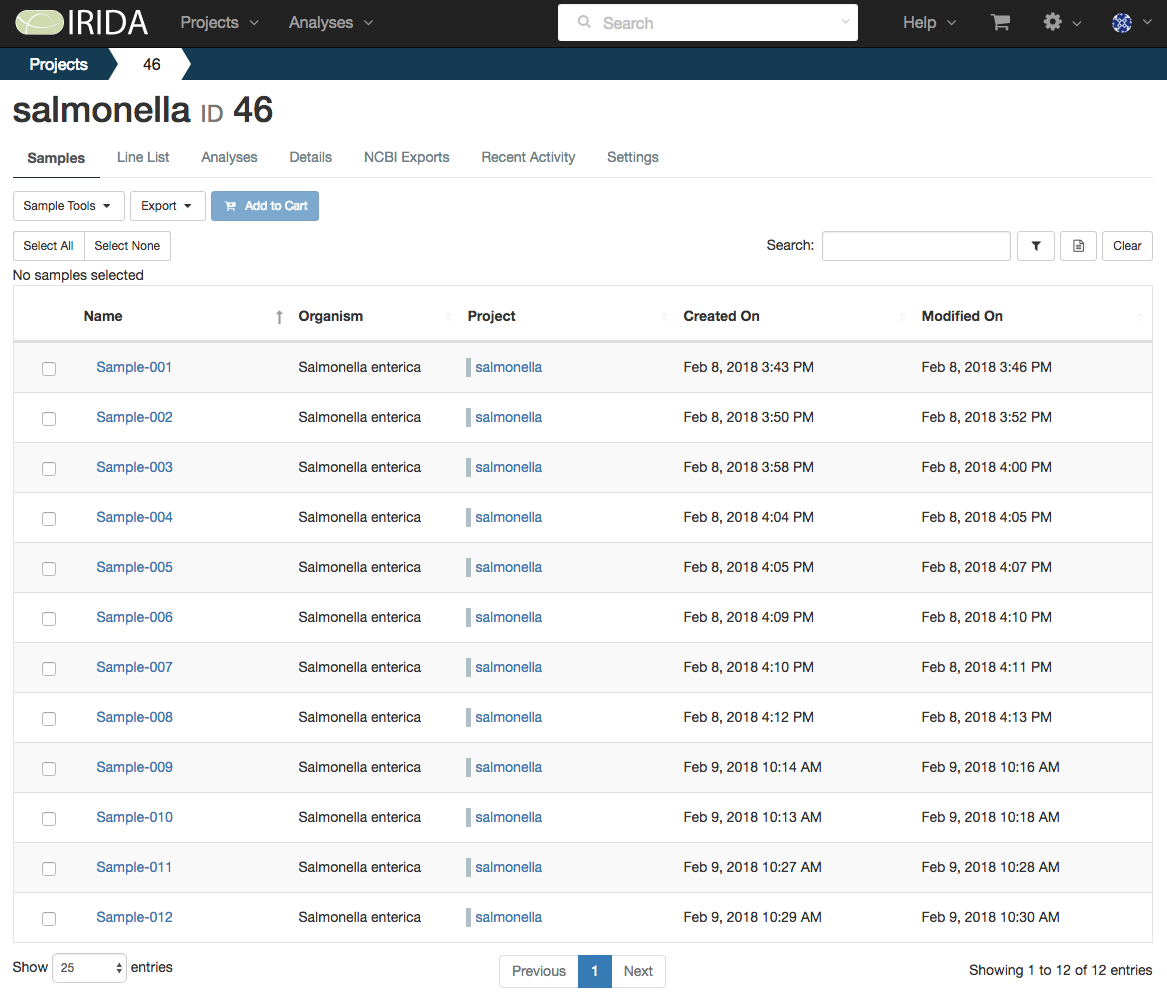 The IRIDA platform offers a fully-featured data management system which enables users to create Sample pages (containing metadata and IDs for tracking throughout the system), which can be stored within Projects. Samples can be used across multiple Projects. Users can also restrict access to Samples and Projects using IRIDA’s permission-based authentication framework. Samples and Projects can also be shared with other users and instances using this same system.
The IRIDA platform offers a fully-featured data management system which enables users to create Sample pages (containing metadata and IDs for tracking throughout the system), which can be stored within Projects. Samples can be used across multiple Projects. Users can also restrict access to Samples and Projects using IRIDA’s permission-based authentication framework. Samples and Projects can also be shared with other users and instances using this same system.
Workflows and Analyses
IRIDA’s sample browser and “shopping cart” system allows users to easily select a collection of samples and sequencing data for analysis, then guides users through selecting a pipeline, choosing appropriate parameters for their sample data, and launching their analysis pipeline. Users can upload sequence reads from Illumina MiSeq sequencing instruments using the MiSeq Uploader, as well as FASTQ files, and assemblies from various sources e.g. public repositories (NCBI). We are also working towards supporting Nanopore data. Raw sequence data can then be curated for quality, and submitted to a number of different workflows for processing and analysis, including:
- The Assembly and Annotation Pipeline which performs de novo or referenced-based assembly and annotates genes and other genomic features. An assembled and annotated genome is generated from reads using Shovill, Prokka and QUAST. Outputs produced by this pipeline include: log files, assembly statistics, the contigs (all contigs, filtered contigs with repeats, filtered contigs without repeats), and annotations from Prokka.
- The Antimicrobial Resistance (AMR) Gene Detection Pipeline, which uses both RGI and StarAMR to identify resistance genes.
- The SNVPhyl Pipeline, which generates single nucleotide variant (SNV) distance matrices and SNV-based phylogenies.
- The MentaLiST Pipeline, which uses k-mer based core genome Multilocus Sequence Typing (cgMLST). This tool genotypes bacterial isolates directly from reads.
- The Salmonella In Silico Typing Resource (SISTR) which performs Salmonella serotyping.
- The RefSeq_Masher Pipeline, which will identify which NCBI RefSeq genomes that most closely match or are contained within your sample sequences.
- The Biohansel Pipeline, which genotypes clonal microbial whole-genome sequencing data.
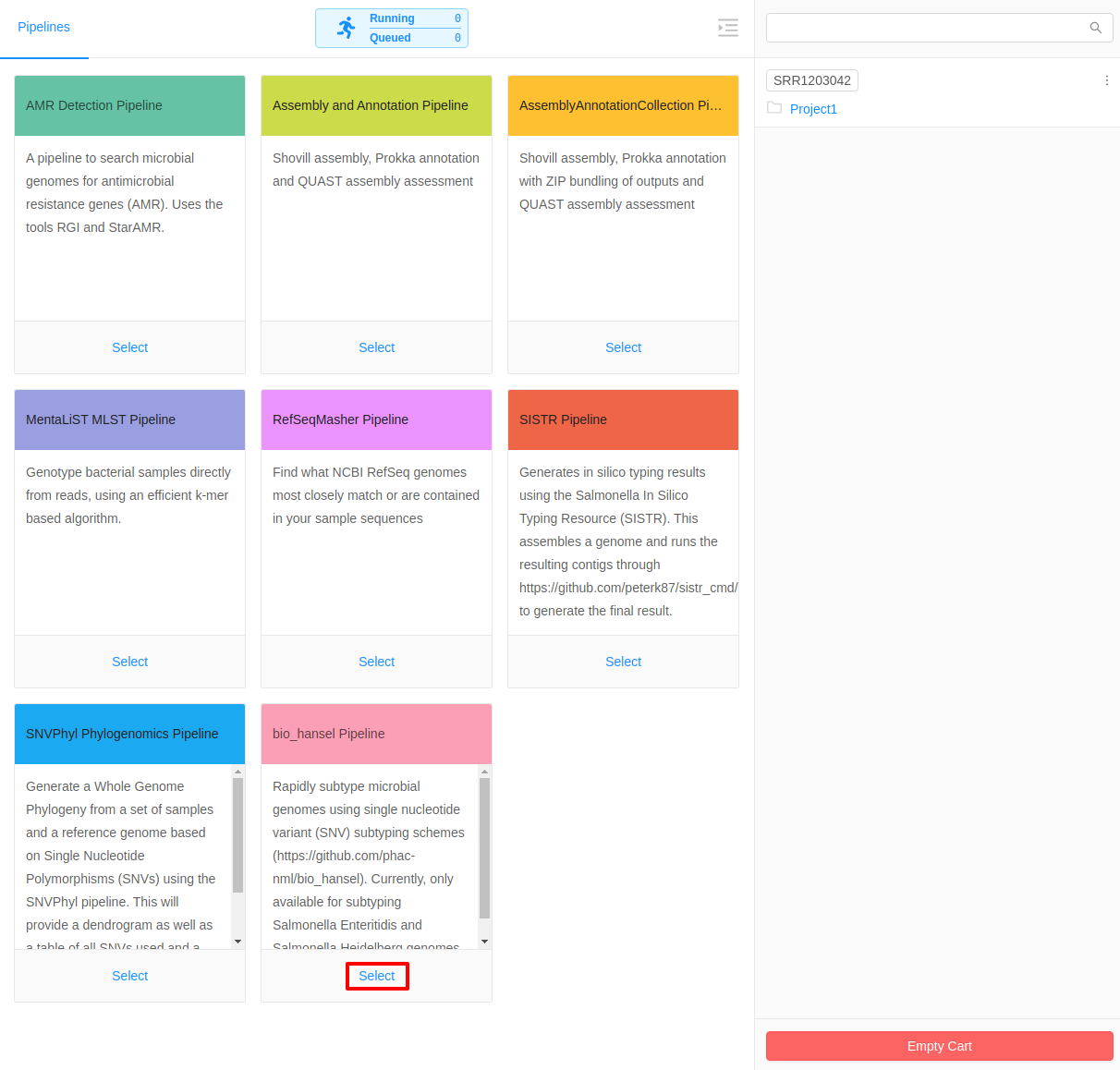 IRIDA offers a variety of processing and analytical pipelines for quality control, assembly, annotation, SNV-based phylogeny construction, cgMLST and Salmonella serotyping.
IRIDA offers a variety of processing and analytical pipelines for quality control, assembly, annotation, SNV-based phylogeny construction, cgMLST and Salmonella serotyping.
While IRIDA aims to package and automate many common analysis activities, we understand that researchers may want to use analysis tools that are not included in the IRIDA platform. To enable this, IRIDA allows developers to write tools to pull data from the IRIDA system for custom analyses. An example is our IRIDA Galaxy export tool. This allows authenticated users to securely pull data from IRIDA into their local Galaxy analysis platform to apply any custom workflows and tools they have installed.
IRIDA also has external connectivity with Applied Maths BioNumerics platform, and NCBI’s Sequence Read Archive.
For more information on writing tools to interact with IRIDA, see our REST API Documentation.
Data Sharing
IRIDA is designed to support collaborative data sharing and analysis. This allows sequencing centres to store and analyze their own data locally while being able to contribute their results to a larger global context. This decentralized model reduces infrastructure requirements and allows smaller centres to work more independently.
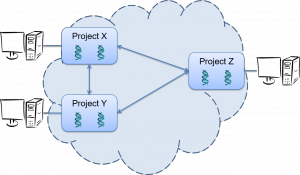 IRIDA administrators can link up their IRIDA systems over the internet and automatically pull project data, sequencing data, and metadata between instances with authentication control via the industry standard OAuth2 framework.
IRIDA administrators can link up their IRIDA systems over the internet and automatically pull project data, sequencing data, and metadata between instances with authentication control via the industry standard OAuth2 framework.
This allows users over multiple IRIDA installations to analyze the same datasets for larger research projects or outbreaks. User role permissions control visibility and editing of content. Sample data and results can be shared between IRIDA instances simply by changing user profile permissions, which can be customized according to data governance guidelines.
Metadata and Visualizations
Metadata can also be stored alongside sequences for describing and tracking samples. Contextual metadata such as specimen type, geographical location, exposures, collection date, case and investigation IDs, can be uploaded through IRIDA’s Metadata Manager uploader, which accepts user-defined spreadsheets (.csv files). Metadata in sample files can be edited once uploaded. Metadata can also be uploaded to sample files without the addition of sequences to facilitate the comparison of epidemiological and laboratory information in line lists. Metadata can be searched, and fields of data can be included or excluded from analysis using customizable filters. Data can also be re-ordered within the line list using the Toggle features.
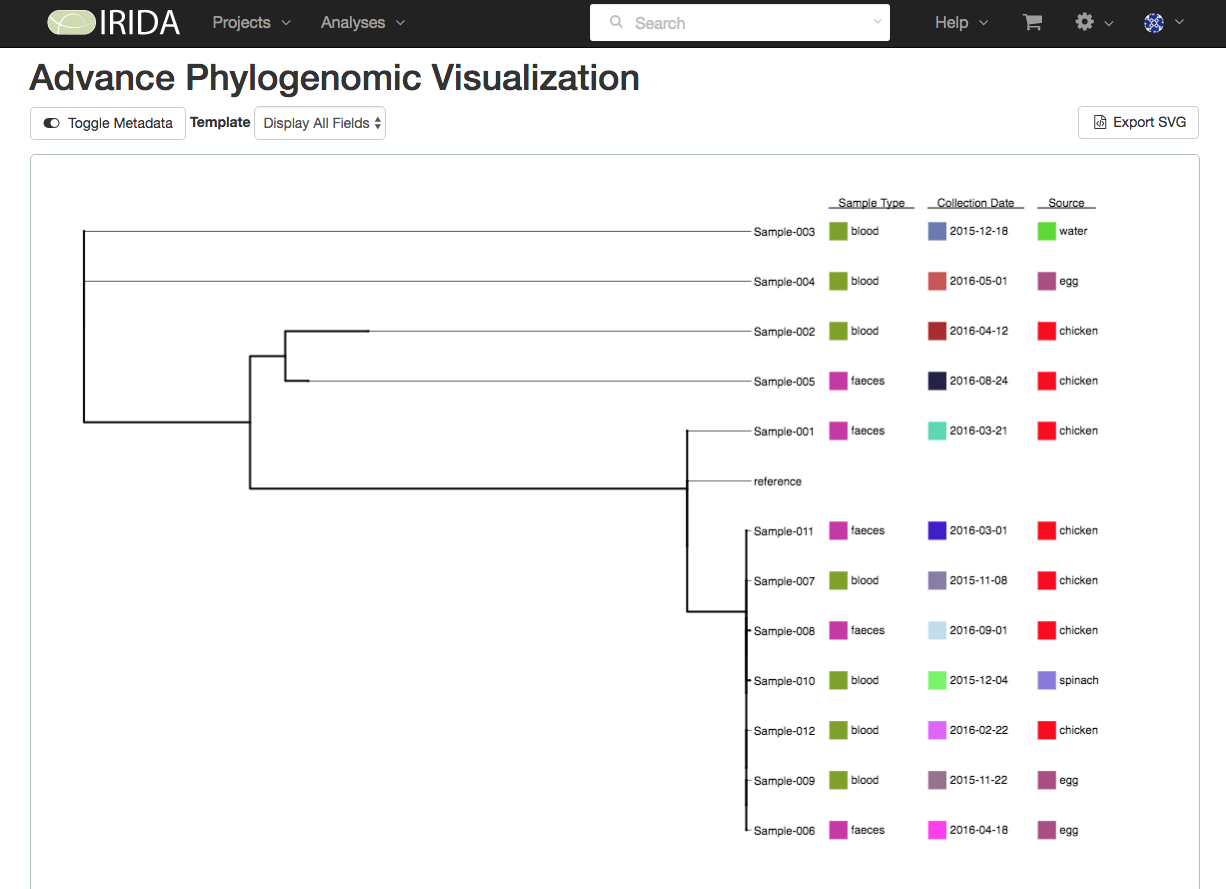 Uploaded metadata can be used to label branches in phylogenomic trees for cluster analysis and decision-making using IRIDA’s Advanced Phylogenomic Visualization tools.
Uploaded metadata can be used to label branches in phylogenomic trees for cluster analysis and decision-making using IRIDA’s Advanced Phylogenomic Visualization tools.
Enhanced Data Harmonization and Integration
IRIDA is the world’s first public health genomics platform to develop and implement ontology to offer standardized terms for describing metadata, which can improve data harmonization and integration between different groups and organizations. Best data stewardship practices encourage the encoding and storage of digital assets such as sequence metadata through the use of community standards like minimum information checklists and ontologies, which better prepare this information for different applications. As such, the IRIDA team has created the Genomic Epidemiology Ontology (GenEpiO) to describe sample details and provenance, as well as clinical, epidemiological, lab, and genomics data and methods.
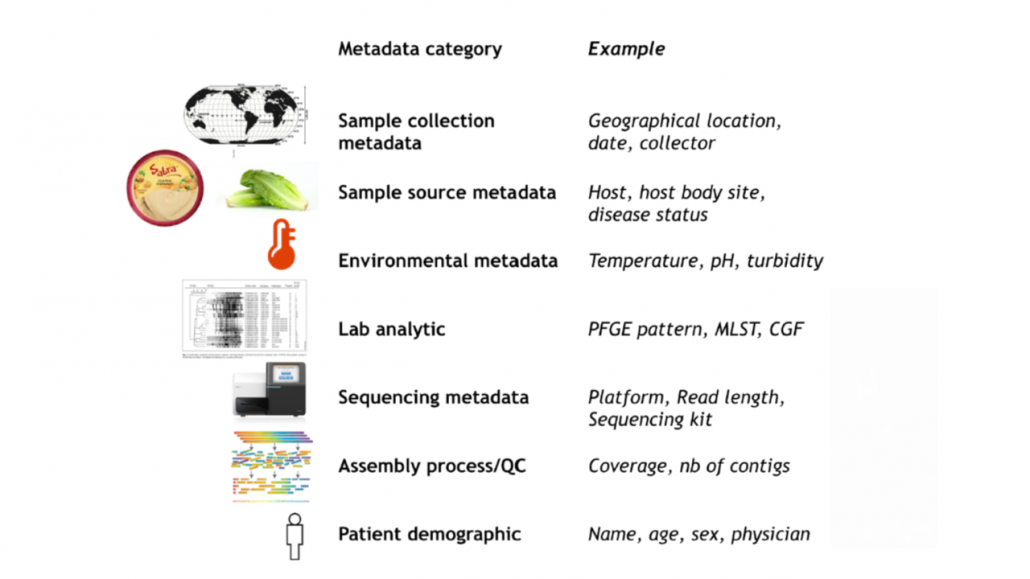 Contextual information required for surveillance and outbreak investigation activities.
Contextual information required for surveillance and outbreak investigation activities.